First steps
ClinicaDL is the deep learning extension of Clinica, an open source python library for neuroimaging preprocessing.
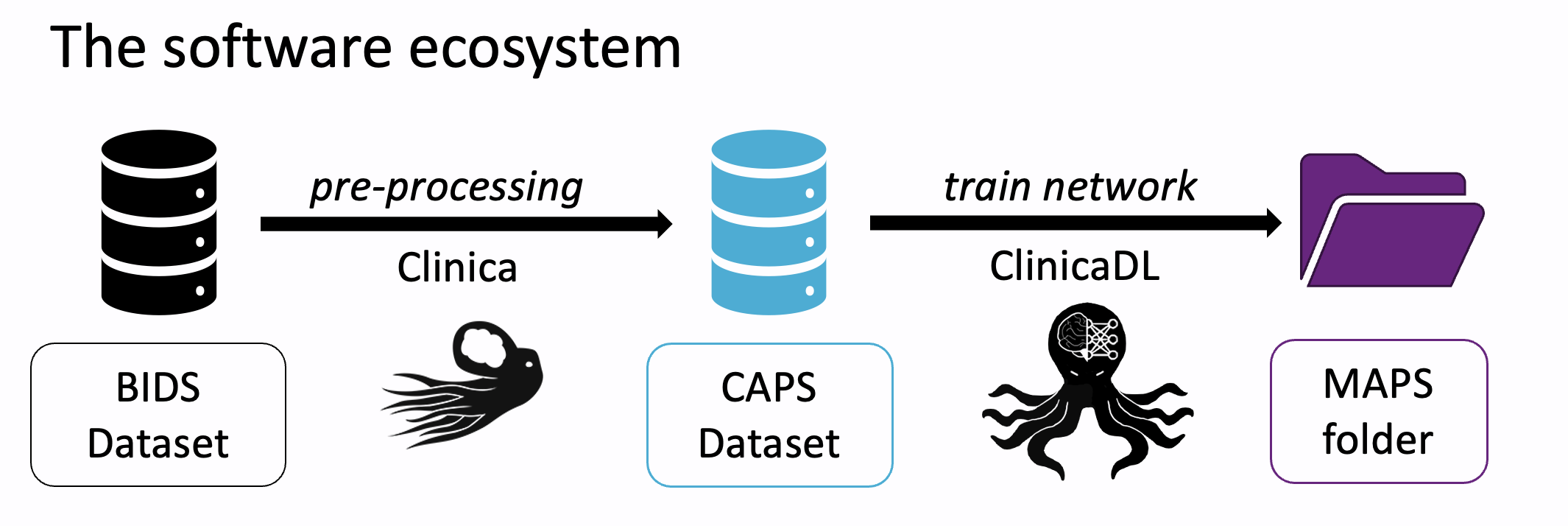
Data formatting¶
Clinica (and so ClinicaDL) relies on raw input data sets formatted with the BIDS (Brain Imaging Data Structure) standard. This standard is largely used in the neuroimaging community, and many data sets are available in this format, or tools were developed to convert them to BIDS.
Clinica preprocesses the images in BIDS format and outputs a CAPS (ClinicA Processed Structure) folder in which preprocessed images are stored. The CAPS format can be seen as an extension of the BIDS.
Finally, ClinicaDL takes as input these preprocessed images to train deep neural networks. To guarantee reproducibility, the training procedure outputs multiple files which are organized according to a specific structure: the MAPS (Model Analysis and Processing Structure).
MAPS definition¶
A MAPS folder contains all the elements obtained during the training and other post-processing procedures applied to a particular deep learning framework. The hierarchy is organized according to the fold, selection metric and data group used.
- fold: In ClinicaDL, a training procedure consists in training one model per train / validation split defined by the validation
procedure. Then the MAPS contains
Nfolders namedsplit-<i>with i between 0 and N-1 containing the best networks obtained for thei-th train / validation split. - selection metric: for each fold, one network is selected per
selection_metricsasked by the user (see Implementation Details for more information). The output folder containing all information linked to a network selected according to a metricmetricis namedbest-<metric>. - data group: Finally, network depending on a fold and a selection metric can be applied to any data group to compute individual results and metrics and interpretability maps. These data groups are defined at the root of the MAPS to ensure that their characteristics are shared across all the models.
train and validation groups
train and validation data groups are automatically created during the training procedure.
These groups can not be changed (overwritten) by the user.
Moreover data of this group is fold dependent, then it can only be loaded and used if a fold number
is specified (contrary to other groups).
prevention of data leakage
The ensemble of all data seen by the network during the training procedure (train + validation data)
is summarized in groups/train+validation.tsv file.
If transfer learning was used, this file also comprises the participants and sessions seen during pretraining.
To prevent data leakage, ClinicaDL prevents the user from creating a data group having a participant ID intersection
with this training data.
An example of a MAPS structure is given below:
<maps_directory>
├── environment.txt
├── split-0
│ ├── best-loss
│ │ ├── model.pth.tar
│ │ ├── train
│ │ │ ├── description.log
│ │ │ ├── train_image_level_metrics.tsv
│ │ │ └── train_image_level_prediction.tsv
│ │ └── validation
│ │ ├── description.log
│ │ ├── validation_image_level_metrics.tsv
│ │ └── validation_image_level_prediction.tsv
│ └── training_logs
│ ├── tensorboard
│ │ ├── train
│ │ └── validation
│ └── training.tsv
├── groups
│ ├── train
│ │ ├── split-0
│ │ │ ├── data.tsv
│ │ │ └── maps.json
│ │ └── split-1
│ │ ├── data.tsv
│ │ └── maps.json
│ ├── train+validation.tsv
│ └── validation
│ ├── split-0
│ │ ├── data.tsv
│ │ └── maps.json
│ └── split-1
│ ├── data.tsv
│ └── maps.json
└── maps.json
The different levels of the MAPS hierarchy are described in the following sections.
Level 1 - validation procedure & groups¶
The first level of the MAPS hierarchy contains the following files:
<maps_directory>
├── environment.txt
├── split-0
├── ...
├── split-<N-1>
├── groups
└── maps.json
environment.txtcontains the description of the environment used for the experiment (output ofpip freeze).maps.jsonis the global configuration file of the MAPS which contains all the information to reproduce the training procedure.split-<i>is a folder containing the result of the training on thei-th split of validation procedure. All possible folds may not be present if the user chose to train only a subset of folds.groupsis a folder in which all the data groups used by the MAPS are defined. This folder has the following file system:groups ├── train+validation.tsv ├── train │ ├── split-0 │ │ ├── data.tsv │ │ └── maps.json │ ├── ... │ └── split-<N-1> │ ├── data.tsv │ └── maps.json ├── validation │ ├── split-0 │ │ ├── data.tsv │ │ └── maps.json │ ├── ... │ └── split-<N-1> │ ├── data.tsv │ └── maps.json └── <data_group> ├── data.tsv └── maps.jsontrain+validation.tsvfile contains all the participant and session IDs used during training (and pretraining if applicable).trainandvalidationdata groups contains one folder per fold in the validation procedure. Then each fold contains the same files than<data_group>.<data_group>contains two files:data.tsvcontaining the list of participant_IDs, session_IDs and the label (if applicable), andmaps.jsonthe configuration file of the group which contains the path to the CAPS directory and a boolean value defining if the group is using a multi-cohort setting.
Level 2 - metrics of selection & training logs¶
The second level corresponds to the content of folder split-<i>. It contains the best models selected during training according to the validation
performance according to a metric metric. It also contains the evolution of all evaluation metrics computed on the training
and validation sets in two different formats:
- tensorboard is a folder containing logs that can be visualized with the command tensorboard --logdir <maps_directory>/split-<i>/training_logs/tensorboard,
- training.tsv is a TSV file.
split-<i>
├── best-<metric>
└── training_logs
├── tensorboard
│ ├── train
│ └── validation
└── training.tsv
Level 3 - model application to data groups¶
The last level corresponds to the content of best-<metric>. In this folder, the weights of the model are available (best_model.pth.tar)
and their application to different data groups can be found:
best-<metric>
├── model.pth.tar
├── train
├── validation
└── <data_group>
train and validation data groups are automatically created as their predictions are computed during the training procedure.
Other groups may exist if predictions and interpretations were computed.
The content of the data group folders depend on the operations performed, then for more information please refer to the corresponding sections.